Inconsistency in evaluation scores
Description
The scores assigned by Salt to jets change depending on the batch size used to process the data.
If a h5 file dumped from the TDD is processed with the default batch size, specific jets will find themselves with scores that do not match what is obtained by processing the same data using a batch size of 1. Or by dumping the individual events in separate h5 files and processing them separately.
Reproducing the issue
Instructions to reproduce the issue follow:
- Download the contents of this directory.
-
185022.h5File for the single event. -
full_stat.h5File for the full sample. -
full_stat_b1.h5File for the full sample, processed with batch size = 1. - The h5 files have been dumped from
mc21_14TeV.601229.PhPy8EG_A14_ttbar_hdamp258p75_SingleLep.deriv.DAOD_FTAG1.e8481_s4149_r14700_r14702_p5799
-
- Open
python3, ensure you haveh5pyinstalled. - Run the following commands:
import h5py
full = h5py.File("full_stat.h5")
full_b1 = h5py.File("full_stat_b1.h5")
full["jets"]["ujets", "cjets", "bjets"][full["jets"]["eventNumber"] == 185716]This will show the following output: 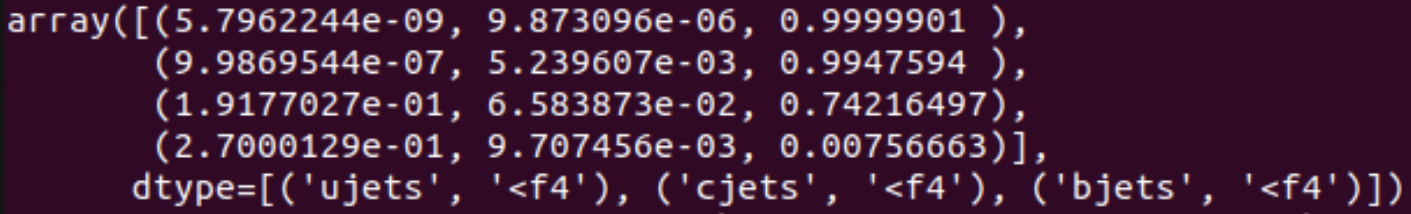
Then run:
full_b1["jets"]["ujets", "cjets", "bjets"][full_b1["jets"]["eventNumber"] == 185716]This will show the following output: 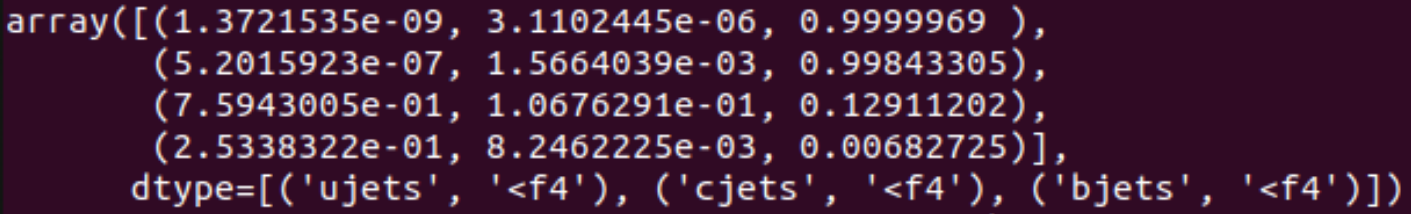
As shown in the image, the scores for the same jets, in the same event, are not compatible between eachother, cross validation with Athena (Athena scores available as GN2HL_px) suggests that the scores given by processing with a batch size of 1 are the correct ones.
However, this does't fix every event, as can be seen by checking with eventNumber == 185022. When processing this event as part of the full stat file, even with the batch sise set to 1, the scores obtained do not match what is produced by dumping a h5 containing only this event and processing that. The results of processing this specific event in a separate file can be obtained with the following Python command:
single = h5py.File("185022.h5")
single["jets"]["ujets", "cjets", "bjets"]Optionally it is possible to run the Salt inference by utilising the 185022.h5 and full_stat.h5 files from the raw/ directory together with the contents of logs/. To do so:
- Edit
logs/GN2_20231219-T092645/config_0_5.yamland replace all occurrences of<your path>with the path to this directory. - Run the Salt evaluation following the instruction on the documentation.
- Run the same python commands as listed above, using the h5 files in the
logs/GN2_20231219-T092645/ckpts/directory.
